- About us
- Research
- Students & Teaching
- Seminars & Events
- Directories
- Booking Rooms & Equipment
- עברית
The Alexander Silberman Institute of Life Science
Research Groups
 Universal Principles of Tissues in Health and Disease The tissues in our body are made up of different types of cells that communicate with each other to perform joint tasks. Tissues differ greatly from each other in the composition of the cells, spatial structure, and function.
In our research we develop computational methods and mathematical models and analyze experimental data to discover the universal principles in intercellular communication that are essential for the proper functioning of the tissue. |
 Microbial ecology and biogeochemistry of the ocean Our mission focuses on unraveling the biomolecular mechanisms employed by microorganisms to control the cycling of elements in the marine environment.
|
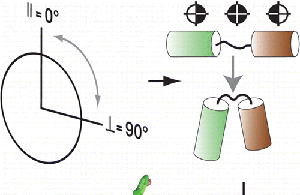 Structural Biology of Membrane Proteins Computational and experimental structural biology of membrane proteins: use and development of novel experimental and computational approaches to elucidate membrane proteins structure. |
|
Cell Biology of Infectious Diseases Membrane traffic in health and disease (endocytosis, exocytosis, transcytosis); Membrane domains; Epithelial cell polarity; Cell biology of enteric bacterial pathogens; Pathogenic bacteria – epithelial host interactions; Inflammatory bowel diseases (Crohn’s disease, ulcerative colitis) |
 Peptide Therapy for Neurodegenerative & Inflammatory Diseases The mechanism of exocytosis; Neurodegeneration: Development of drugs for Parkinson’s and Alzheimer’s disorders. |
 How Do Plants Degrade Cellular Components for Energy Production We are interested in understanding how plants recycle cellular components when disposing of unwanted proteins and organelles for energy production. This occurs during the development of non-photosynthetic tissues and under stress conditions in which photosynthesis is not functioning well. |
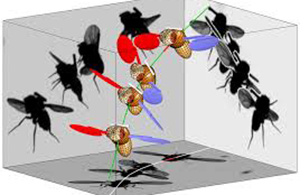 Micro Flight Lab Understanding the mechanisms of flight control in insects; bio-mimetic robotics |
 Basic & Applied Aspects of Environmental Microbiology Microbial ecology of extreme environments, with a focus on the microbial populations of salt-excreting plants; Microbial water quality; Genetically engineered microbial sensors for environmental monitoring, with an emphasis on toxicity and genotoxicity assessment and on the detection of specific classes of compounds, including trace pharmaceuticals and explosives. |
|
Genetic Regulation of CNS Development & Neurodegenerative Diseases Developmental Neuro-Genetics: the study of bHLH transcription factors regulating embryonic development, focusing on the nervous system. Characterization of the role and function of the transcription factors at the cellular and whole organism levels, utilizing chick embryos and knockout and transgenic mice. Revealing their molecular mode of action at various levels (transcription regulation of target genes, molecular interactions: DNA-protein, protein-protein etc.). We also study the involvement of transcription factors in human diseases (developmental, inherited and cancer). |
 Human Embryonic Stem Cells Differentiation and genetic manipulation of human embryonic stem cells ; Human genetic disorder and human embryonic stem cells ; Genomic analysis of human development ; Oncogenic properties of stem cells |
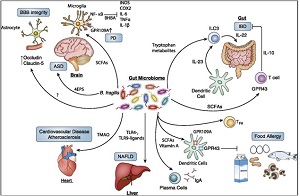 The Gut-Brain Axis The past decade has witnessed a paradigm shift in brain research. A transition from a dogmatic brain-focused approach toward a holistic conception of health that integrates key signaling hubs of the human body—such as the gut and its microbial populations, the peripheral immune system, and other mucosal barrier surfaces—is increasingly acknowledged as necessary to understand and cure neurological disorders, such as Alzheimer’s disease, stroke and brain tumors. We investigate the physiological connections between the digestive, immune and nervous systems in a holistic approach, allowing better understanding of neurological diseases in the context of aging. We study how processes which take place in the gut, affect brain functions in health and disease, and looking at the whole-organism level to identify novel targets to treat brain pathologies. |
 Sociobiology, Chronobiology & Physiology of Bees We study the evolution and mechanisms underlying sociality and social behavior, using bees as a model. Our main research focuses are:
|
 Systems genetics of green microalgae – from fundamental cell biology to translational applications
Building on the versatile and magnificent world of green microalgae, we investigate basic principles of plant cell biology and translate our understandings into innovative technologies.
We focus on the chloroplast, the plant’s major energy producer, and seek to advance our mechanistic understanding of the biogenesis and protein quality control of this critical plant organelle in steady-state and stress conditions.
Our uniqueness is in the toolkit. In order to gain true mechanistic understanding, we develop clever genetic tools, forward and reverse genetic screens and fluorescence-based large-scale imaging approaches at a single-cell resolution, in the powerful Chlamydomonas reinhardtii.
Alongside our fundamental science goals, we seek to pioneer agritech and foodtech solutions. This includes engineering algae for multiple functions, turning them into a groundbreaking production platform for nutrients and proteins and paving the way for plants that are highly resilient to global warming and can better sustain life on our planet into the next millennia.
|
 Mechanobiology of Stem Cells and Nucleus Biophysics Cell-matrix interactions in directing cell-fate decision making, Remodeling of the nuclear matrix of lamins and chromatin |
 Ancient DNA & Human Evolution The ability to sequence DNA from bones and teeth opened the opportunity to read the genetic makeup of ancient populations like Neanderthals and Mammoths. The work in my lab focuses in identifying the genetic changes that made us humans, in studying the epigenetics of ancient populations, and in general understanding evolutionary and historical processes. |
 Developmental & Evolutionary Biology Studying arthropod evolution using different sources of information: Comparative embryology, comparative genomics, paleontology and systematics. |
 Experience-Dependent Plasticity Neuroscience, molecular mechanisms of learning and memory, drug addiction, feeding disorders, experience-dependent plasticity, neural circuit adaptations, synaptic physiology and plasticity, transcriptional regulatory networks, non-coding RNA. |
 Pain & Anesthesia Research Laboratory Plasticity in the somatosensory system in mammals. Neural mechanisms whereby injury provokes sensory dysfunction and chronic pain. Pathophysiology of injured nerve and abnormal impulse generation. Synaptic reorganization in the brain and spinal cord after peripheral nerve injury. Regeneration and collateral sprouting of injured nerve fibers. Animal models of chronic pain states and pain relief. Heritability of chronic pain. Neural mechanisms of loss of pain response and of consciousness in general anesthesia. |
 Epigenetics, Chromatin & Cancer Epigenetic regulation of gene expression. Cancer epigenetics. |
 Developmental Mechanisms of inflammatory Diseases & Cancer Development of constitutively active MAP/stress kinases for studies of the etiology cancer and inflammation. Revealing the mechanism of gene expression under stress in yeast and mammalian cells. Focusing on Gcn4, Hsf1 and Msn2/4. Analysis expression of stress-related genes in oncogenically-transformed cells. |
 Marine ecosystems in a changing world The marine environment is challenged by global and local environmental changes. Our laboratory is engaged in understanding the processes that shape marine ecosystems under scenarios of environmental change. From observations at sea, to controlled aquarium systems and laboratory experiments, our group works to understand processes that will shape the marine environment in the years to come. |
 Terrestrial microbial ecology and plant-microbe interactions
The lab’s goal is to promote a systems-level understanding of the rules that govern the assembly and maintenance of a functionally robust plant microbiome and its effects on plant health.
We aim to understand how the ecological context, in particular microbe-microbe interactions, determines microbiome assembly and the resulting plant phenotypes.
|
 Marine Microbial Eco-Physiology Biological oceanography; Marine phytoplankton ecology and cell biology; Population dynamics; Life cycle strategies. My lab focus on the investigation of the main ecological and cellular differentiation between life cycle phases, understanding the role of each phase and the interplay with other organisms in the oceans as well as on the search for triggers regulating life phase transitions. |
 Developmental Biology, Regeneration and Evolution of the Sea-Anemone Nematostella A main subject in the lab is the gene expression changes underlying the process of whole-body regeneration (the remarkable ability of some animals to form their body again after dissection into pieces) in the sea anemone Nematostella vectensis. We are analyzing the molecular genetics at the base of this amazing regenerative ability and compare this process to embryonic development. In our research we have discovered a little-known gene family, which has a role in regeneration but is also notorious for being involved in the metastasis of cells in many aggressive cancers in man. |
|
Biological Oceanography & Coral Reef Ecology Benthic-pelagic coupling. Predator-prey interactions in the marine environment. Biomechanics and effects of flow on corals, invertebrates and fish. |
 Early Events in Cancer Development Cancer genetics, Early events in cancer development, Cell cycle, Genomic stability, The Cellular response to DNA damage , Cell cycle checkpoints , DNA repair |
 Circadian Rhythms in Plants Circadian rhythms in plants:
|
 Ecological genomics & evolutionary modeling We are interested in dynamics of eco-evolutionary processes and how they can be read from studying genomic data. We model and analyze population genomic datasets of humans and other species, with applications to conservation biology and paleogenomics. We are also interested in an emerging species control technology called 'gene drives', and are using models to understand the ecological aspects of its potential deployment in the wild. |
 Experimental Biology of Vertebrate Aging & Age-Related Diseases To address a major challenge in aging research, the lack of short-lived vertebrate genetic model, I have developed a comprehensive genetic platform for rapid exploration of aging and disease in the shortest-lived vertebrate model, the African turquoise killifish.
Experimental biology of vertebrate aging and age-related diseases using the short-lived African turquoise killifish; The genetic basis behind the outstanding diversity of lifespan between different animals; Genetic engineering and live imaging of age-related traits in multiple complex organs. |
|
Functional Ecology of Food-Web Dynamics Ecological and evolutionary aspects of predator-prey interactions, stress ecology, inducible defenses, above-below ground interactions, ecological-traps, nutritional ecology, individual syndrome, ecology and evolution of conspicuous anti-predator colors and patterns, herpetology and especially lizard ecology, desert ecology, ecosystem conservation and restoration. |
 Genetics, Molecular Biology and Genetic Engineering in Plants Synthesis of carotenoid pigments in plants; Genetic regulation of natural products in the fruit; Gene cloning from tomato and their molecular analysis; Genome editing in tomato; Biotechnology and genetic engineering of natural compounds in tomato; Regulatory mechanisms of the hormone ABA in the plant. |
|
Cell Biology of Malignant Cells The relation between cell adhesiveness and growth control in malignant lymphoma cells. Molecular mechanisms involved in the selection of non-malignant variant cells from malignant cell populations. Different approaches to reversal of multi-drug resistance of malignant cells. Growth regulation of malignant lymphoma. Metastasis of malignant lymphoma. |
 Motor Control of the Flexible Arms of the Ooctopus - Inspiration for Rrobotics
|
|
Visual Systems Vision: information processing in the mammalian and primate visual pathway. Computational neuroscience. Perception, learning and conciousness. |
 Biodiversity & Nature Conservation Plant population and community ecology. Plant demography; vegetation dynamics; succession. Landscape ecology, geographical information systems (GIS); conservation. |
 Computational Structural Biology Structural biology of large protein complexes. Mass-spectrometry. Computational structural modeling based on multiple information sources. Protein structure prediction. |
 Ecology - Modeling and Theory We study how can some ecological communities harbor hundreds of relatively similar species in a small area, while others have just a few? Why do ecological communities change in space and time, and what can these changes teach us? And how can we use data science and models to study these extremely complex systems? |
 Genomic Instability in Human Cancer & Genetic Diseases
|
|
Studying photosynthetic life in a dynamically changing environment The Keren research group works on the interaction of photosynthetic organisms with natural environmental conditions. Our lab is currently working on two aspects of this problem (a) the transport, storage and assembly of transition metals into photosynthetic catalysts (b) Elucidating the biophysical principles of energy and electron transport in photosynthetic systems. |
 Modeling Ecological & Evolutionary Dynamics |
 Mechanism & Regulation of RNA Polymerase II Transcription
Our research is directed towards the mechanism and regulation of RNA polymerase II transcription. We seek to reconstitute the entire process from promoter chromatin remodeling to transcript synthesis with pure proteins and nucleic acids, to solve the structures of the proteins, and to elucidate their functional interactions.
|
 Single-Molecule Biology
|
 Signal Transduction Therapy Signal transduction, Signal transduction therapy,Cancer Research, Medicinal Chemistry. |
 Neeuroscience & Computational Biology The dynamic processes of nerve terminals and the molecular aspects of synapse functioning. Control of exocytosis in regulated secretory systems. Differentiation and neurotransmitter phenotype acquisition in neurons, a study by proteomics and genomics approaches. Bioinformatics. Large scale studies of biological sequences and their global organization. Proteomics. Structural genomics. |
 The interactions Between Pathogens, the Host and the Gut Microbiota
We explore molecular mechanisms that control the balance between bacterial pathogens, the gut microbiota, and the host.
Our overarching goals are:
Our research strategy is to address these fundamental biological questions by taking a triangulated approach:
|
|
Stuctural Biology of Proteins Structural determination of biologically related macromolecules via X-ray crystallographic techniques. Ligand recognition, initial signaling events, and the role of dimer/oligomer orientation in cytokine receptor systems. Structural studies of avidin/streptavidin - biotin high affinity systems. Structural based drug design and optimization. Combinatorial approaches in drug discovery. |
 Decision Making Research Decision-making and reinforcement learning, Reinforcement learning and cellular plasticity, The neuronal mechanisms underlying contraction bias, the functional architecture of the cerebellar cortex. |
|
Neural Coding Neural and dendritic computation and coding, Neuronal noise, neural basis of vocal communication, information theory analysis of neural communication. |
|
Epigenetics, Stem Cells and Neurons Epigenetics in stem cells, neuronal differentiation and neurons; Modeling neurodegenerative diseases using pluripotent stem cells; Paleo-epigenetics: reconstructing methylation in ancient DNA; Live imaging of nuclear dynamics in stem cell differentiation; Signaling pathways and molecular networks in stem cells and cancer |
 Tissue Engineering Genes coding for factors involved in control of differentiation and development. Control of proliferation and differentiation in adult tissues. Epithelial mesenchymal interactions. Gene therapy, angiogenesis, ageing. Differentiation of stem cells. Cell therapy. |
 Neuronal & Circuit Plasticity
What are the mechanisms of neuronal plasticity in the brain? Functional and structural plasticity during perceptual learning. What changes in the brain underlie social learning, and maternal behavior. The auditory system – development, learning and function.
|
 Molecular Evolution The roles of microRNAs in cnidarian development; Evolution of sea anemone (and other strange animal) toxins; Evolution of voltage-gated sodium channels |
 Translation regulation by RNA modifications in hematopoietic stem cells and disease
Our goal is to understand the regulatory role of the ribosome in the central dogma of gene expression.
We explore molecular mechanisms of translation-regulation, focusing on the role of RNA modifications in translational control during normal hematopoietic development, leukemia initiation, and viral infection.
In our studies we utilize many molecular techniques, such as ribosome profiling, RNA modification sequencing and CRISPR-based screens. We combine these with ex-vivo stem cell research and in-vivo models by bone marrow transplantation, and leukemia xenograft. Using this diverse tool box we aim to reveal the centrality of cell-type-specific translation for the establishment of cell identity.
|
|
Liver Tissue Engineering & Metabolism 1. Metabolic programing: nuclear reactors-based control of metabolism 2. Human embryonic stem cell differentiation to hepatocytes 3. Liver tissue engineering and liver regeneration 4. Hepatitis C Virus (HCV) infection |
 Movement Ecology Theoretical and applied population and community ecology, and aspects of movement ecology, in particular. Ecology and evolution of dispersal using an integrated theoretical-empirical approach, spatiotemporal population dynamics, Mechanistic models. Gene flow; frugivory plant-animal interactions, bird migration, ornithology, rarity, macroecology, conservation biology. |
 Structure & Function of Photosynthetic Proteins The structure-function relationships of membrane proteins mainly of the photosynthetic apparatus. Chloroplast development and light regulated expression of chlorophyll-protein complexes. |
 The function of plant specialized metabolites Plants synthesize an amazing diversity of specialized metabolites, estimated between 200,000 to 1,000,000 metabolites in the plant kingdom and ~3,000 in each individual species. Despite a growing understanding of biosynthetic mechanisms of many specialized metabolites, the in-planta function of the vast majority remains unknow. We are interested in examining the functions of specialized metabolites in the plants that synthesize them naturally, with an emphasis on protection from herbivory, and functions in a natural setting. In such a setting, we may examine complex interactions including several trophic levels and discover interactions that we didn’t previously hypothesize.
|
 Auditory Neurophysiology Responses of neurons in the auditory system to complex sounds |
 Steroid Endocrinology
|
 Plant Phenotypic Response The main aims of our research are designed to investigate the nature of nuclear-organellar interactions, which link mitochondrial genome status and expression and plant phenotypic response. |
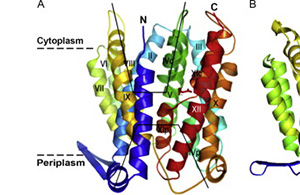 Molecular & Structural Biology of Membrane Proteins Molecular membrane biology, structure biology, bioenergetics, biochemistry, biophysics. |
 mRNA biology in differentiation and early embryogenesis
We study how posttranscriptional regulation of mRNA molecules underlies embryonic development. We investigate different aspects of RNA biology (stability, localization, processing etc.). By answering these questions at the context of development, we aim to uncover general principles of post-transcriptional RNA regulation, that apply to many other biological systems.
We base our work at the intersect of computational and experimental biology, using the zebrafish model system.
|
 Epigenomics Chromatin biology, Epigenomic mechanisms, Cellular differentiation, Cellular heterogeneity, single cell technology |
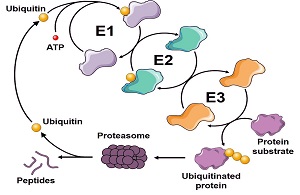 Quality control-associated proteolysis by the ubiquitin-proteasome system High throughput analysis of signals that trigger misfolded protein elimination and the contribution of cellular chaperones.
|
 Protein Homeostasis & Stress Response System biology of redox regulation, intrinsically disordered chaperones, computational and experimental biology,proteomics and mass spectrometry. |
 Neural Conduction Experimental and theoretical neurobiology. Computational neuroscience; the neuron as a complicated computing element; individual nerve cells as information processing devices. Time coding by neurons. |
 Protein Engineering: Modulation of Protein-Protein Interactions Development of computational method for redesign of protein-protein interfaces for altered affinity and binding specificity. the projects include redesign of calmodulin-target interactions, AChE-toxin interactions, Ras-effector interactions, and prion protein interactions. |
 Genetics of Neurodevelopmental Disorders Genetics of autism and schizophrenia; The role of chromatin regulators in brain development and function; Genetics of the variation in gene expression |
 Molecular Genetics of Budding Yeast Molecular genetics in yeast: meiosis, cell cycle, chromosome pairing, segregation and recombination in meiosis. Instability in the human, mouse and yeast genomes. DNA repair. |
 Cholinergic Signaling in Health and Disease Stress responses. Antisense technology. Acetylcholinesterase biology. Molecular neurobiology. Messenger RNA studies |
 Structure and Function of the Nuclear Pre-mRNA Processing Machine Structure and function of the nuclear pre-mRNA processing machine. Pre-mRNA splicing. Pre-mRNA processing. Regulation of gene expression in eukaryotes. Assembly and structural studies of biological systems. Autoantibodies against nuclear RNPs-probes for inflammatory myopathies and RNA processing. Physical biochemistry. Electron microscopy. |
 Structural and Functional Studies of Hepaciviruses
Hepatitis C virus (HCV) is a major health burden that infects 1-2% of the world population and leads to ~500,000 deaths and an estimated 1.5-2 million new infections annually.
Direct-acting antivirals effectively treat HCV infection, yet face major issues making the development of additional therapeutics and an effective vaccine critical to prevent HCV spread.
Our research objectives are:
To achieve these objectives, we utilize structural (X-ray crystallography), biochemical and functional tools. A deep understanding of the interaction between HCV and the host cells and will be use to facilitate structural-based design of novel therapeutics and subunit vaccine immunogens for better controlling of HCV spread.
|
 Telomeres & Telomerase The three-dimensional structure, function and regulation of telomeres and telomerase; Human genetic diseases caused by telomere dysfunction. |
 Germ Cells Development In our group we study different aspects of the biology of fertility. We focus on the study of the way oocytes develop, and what cause their rapid aging. Research students in the lab use a variety of methods such as:
|
|
Neurobiology of the Cerebellum Cellular physiology of central neurons. Subthreshold oscillations, resonance frequencies and rhythm generation in mammalian central nervous systems. |
|
Systems Biology & Neurogenetics As a Systems Biology lab we combine experiments and theory to understand the design principles of biological networks: 1) Computation and functional dynamics in neural circuits with a single neuron resolution. 2) Plasticity in neural networks (Learning and memory, aging, neurodegenerative diseases) 3) Phenotypic variability: from gene expression and neural activity to behavior 4) Evolutionary and functional genomics: Transcription networks and the relationship between gene expression and genome structure. |
 Visual Perception and its Neural Correlates Functional imaging of the human visual system. Coordinate transformations in the visual system. Active vision: the interplay between perception, attention, and eye movements. Interactions between vision and haptics. The human body image. Vision and action: the mirror system. Plasticity in the visual system following blindness. |
