- About us
- Research
- Students & Teaching
- Seminars & Events
- Directories
- Booking Rooms & Equipment
- עברית
Home » Deciphering the architectures of Nsp1, Nsp2, and Nucleocapsid proteins from SARS-CoV-2
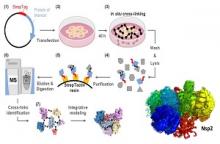
Atomic structures of several proteins from the coronavirus family are still partial or unavailable. A possible reason for this gap is the instability of these proteins outside of the cellular context, thereby prompting the use of in-cell approaches. In situ cross-linking and mass spectrometry (in situ CLMS) can provide information on the structures of such proteins as they occur in the intact cell.
Prof. linial, Dr. Schneidman and Dr. Kalisman applied targeted in situ CLMS to structurally probe Nsp1, Nsp2, and Nucleocapsid (N) proteins from SARS-CoV-2, and obtained cross-link sets with an average density of one cross-link per twenty residues. They then employed integrative modeling that computationally combined the cross-linking data with domain structures to determine full-length atomic models.
For the Nsp2, the cross-links report on a complex topology with long-range interactions. Integrative modeling with structural prediction of individual domains by the AlphaFold2 system allowed us to generate a single consistent all-atom model of the full-length Nsp2. The model reveals three putative metal binding sites, and suggests a role for Nsp2 in zinc regulation within the replication-transcription complex.
For the N protein, we identified multiple intra- and inter-domain cross-links. Our integrative model of the N dimer demonstrates that it can accommodate three single RNA strands simultaneously, both stereochemically and electrostatically.
For the Nsp1, cross-links with the 40S ribosome were highly consistent with recent cryo-EM structures.
These results highlight the importance of cellular context for the structural probing of recalcitrant proteins and demonstrate the effectiveness of targeted in situ CLMS and integrative modeling.